About Hoxa
There are many variations of passages of Lorem Ipsum available but the majority have suffered alteration in some form by injected humour or randomised words which dont look even believable genera on the Internet tend to repeat predefined chunks necessary making Internet.Diffrent Websites
Get in Touch
- 2901 Marmora Road, Glassgow,
Seattle, WA 98122-1090 - (088) -234 -456 -7890
- (088) -234 -456 -7890
- info@yourdomain.com
- http://domainname.com
- marta.filizola@mssm.edu
- + 1 212-659-8690
Mechanistic Insights into GPCR Dimerization/Oligomerization
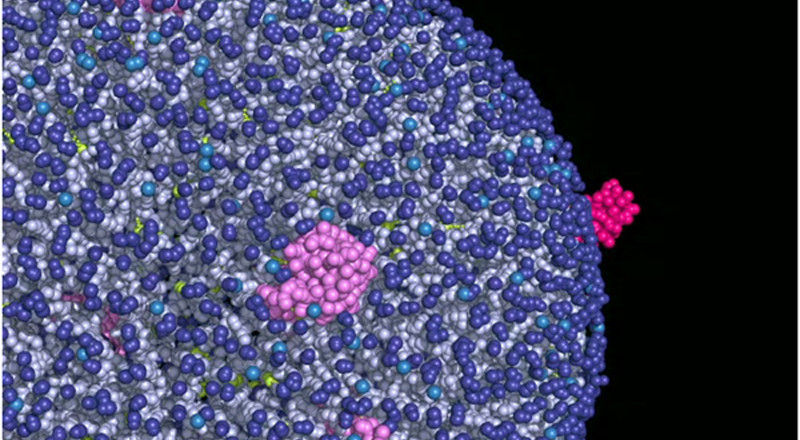
Compelling evidence that models of GPCR signaling must consider oligomeric assemblies rather than isolated monomers started to appear in the literature during my postdoctoral training. Many of the published studies described (i) effects resulting from activating one GPCR in the presence of another and (ii) modulation of the activity of one receptor using ligands targeting another one. Whether these effects resulted from downstream crosstalk or from differential signaling of a receptor complex has so far been difficult to ascertain for the largest subfamily A of GPCRs. My lab has been hard at work in the area of GPCR oligomerization, contributing to their recognition, nomenclature, storing, structural models, and estimates of relative stability. We are committed to providing a rigorous mechanistic insight into the spatio-temporal organization of GPCRs in living cells at a level of molecular detail that is unattainable using current experimental techniques alone, but is required for an ultimate understanding of the role of GPCR oligomerization in receptor function.
Representative Publications
Pin, J.-P., Neubig, R., Bouvier, M., Devi, L., Filizola, M., Javitch, J.A., Lohse, M.J., Milligan, G., Palczewski, K., Parmentier, M., Spedding, M. “International Union of Basic and Clinical Pharmacology. LXVII. Recommendations for the Recognition and Nomenclature of G Protein-Coupled Receptor Heteromultimers.” Pharmacological Reviews (2007) 59:1-9.
Khelashvili, G., Dorff, K., Shan, J., Camacho-Artacho, M., Skrabanek, L., Vroling, B., Bouvier, M., Devi, L., George, S.R., Javitch, J.A.,Lohse, M.J., Milligan, G., Neubig, R., Palczewski, K., Parmentier, M., Pin, J.-P., Vriend, G., Campagne, F., Filizola, M. “GPCR-OKB: A database for G protein-coupled receptor oligomers.” Bioinformatics (2010) 26(14):1804-1805.
Johnston, J.M., Wang, H., Provasi, D., Filizola, M. “Assessing the Relative Stability of Dimer Interfaces in G Protein-Coupled Receptors.” PLOS Computational Biology (2012) 8(8): e1002649.
Provasi, D., Boz, M.B., Johnston, J.M., Filizola, M. “Preferred Supramolecular Organization and Dimer Interfaces of Opioid Receptors from Simulated Self-Association” (2015) PLOS Computational Biology Mar 30;11(3):e1004148.
Marino, K., Prada-Gracia, D., Provasi, D., Filizola, M. “Impact of Lipid Composition and Receptor Conformation on the Spatio-Temporal Organization of mu-Opioid Receptors in a Multi-component Plasma Membrane Model” (2016) PLOS Computational Biology 12(12):e1005240
Meral, D., Provasi, D., Prada-Gracia, D., Möller, J., Marino, K., Lohse, M.J., and Filizola, M. “Molecular details of dimerization kinetics reveal negligible populations of transient µ-opioid receptor homodimers at physiological concentrations” (2018) Scientific Reports 8(1):7705
Walsh, S., Mathiasen, S., Christensen, S.M., Fay, J.F., King, C., Provasi, D., Borrero, E., Rasmussen, S.G.F., Fung, J.J., Filizola, M., Hristova, K., Kobilka, B., Farrens, D.L., Stamou, D. “Single proteoliposome high content analysis reveals differences in the homo-oligomerization of GPCRs” (2018) Biophysical Journal 115(2):300-312.
Thibado, J.K., Tano, J.Y., Lee, J. Salas-Estrada, L., Provasi, D., Strauss, A., Ribeiro, J.M.L., Xiang, G., Broichhagen, J., Filizola, M., Lohse, M.L., Levitz, J., “Differences in interactions between transmembrane domains tune the activation of metabotropic glutamate receptors” eLife (2021) Apr 21;10:e67027.
 Creative Style
Creative Style Portfolio Style
Portfolio Style One Page Style
One Page Style Landing Page
Landing Page