About Hoxa
There are many variations of passages of Lorem Ipsum available but the majority have suffered alteration in some form by injected humour or randomised words which dont look even believable genera on the Internet tend to repeat predefined chunks necessary making Internet.Diffrent Websites
Get in Touch
- 2901 Marmora Road, Glassgow,
Seattle, WA 98122-1090 - (088) -234 -456 -7890
- (088) -234 -456 -7890
- info@yourdomain.com
- http://domainname.com
- marta.filizola@mssm.edu
- + 1 212-659-8690
Integrative and Information-Driven Modeling of Biomolecular Complexes
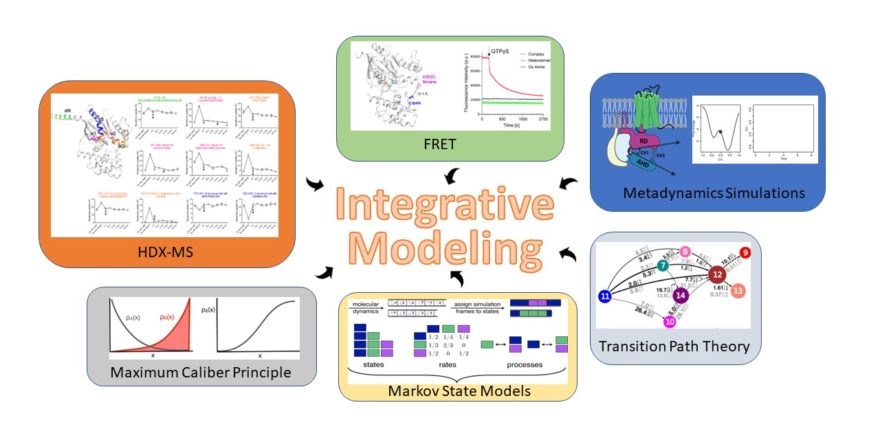
The increased prominence of biomolecular structures solved by cryo-electron microscopy (cryo-EM), coupled with the advancements in proteomics, biophysical, and computational approaches, has recently propelled the popularity of integrative, information-driven computational approaches for modeling the structure and dynamics of biomolecular complexes. In our research, we are focusing on exploring the potential of a novel Bayesian integrative modeling framework that combines computational data from metadynamics, maximum caliber principle, Markov state models, transition path theory, and transfer entropy analysis with experimental data from various techniques, including – but not limited to – pulsed hydrogen deuterium exchange mass spectrometry (HDX-MS), tryptophane-induced quenching (TrIQ), single-molecule fluorescence resonance energy transfer (sm-FRET), double electron-electron resonance (DEER), and cryo-EM to provide a structural context to the kinetics and allostery of G protein-coupled receptor-mediated transducer activation.
Representative Publications
Meral, D. , Provasi, D., Filizola, M. “An Efficient Strategy to Estimate Thermodynamics and Kinetics of G Protein-Coupled Receptor Activation Using Metadynamics and Maximum Caliber” bioRxiv; https://doi.org/10.1101/367888; (2018) Journal of Chemical Physics 149(22):224101 [PMID: 30553249].
Salas-Estrada, L., Fiorillo, B., Filizola, M. “Metadynamics Simulations Leveraged by Statistical Analyses and Artificial Intelligence-Based Tools to Inform the Discovery of G Protein-Coupled Receptor Ligands” (2022) Frontiers in Endocrinology, Section Cellular Endocrinology; 13:1099715. doi: 10.3389/fendo.2022.1099715 [PMID: 36619585]
Ahn, D., Provasi, D., Duc, N.M., Xu, J., Salas-Estrada, L., Spasic,, Yun, M.W., Kang, J., Gim, D., Lee, J., Du, Y., Filizola, M., Chung, K.Y. “Gαs slow conformational transition upon GTP binding and a novel Gαs regulator” bioRxiv (2022) doi: https://doi.org/10.1101/2022.10.10.511514; (2023); iScience Apr 8;26(5):106603. doi: 10.1016/j.isci.2023.106603. [PMID: 37128611 ]
 Creative Style
Creative Style Portfolio Style
Portfolio Style One Page Style
One Page Style Landing Page
Landing Page