About Hoxa
There are many variations of passages of Lorem Ipsum available but the majority have suffered alteration in some form by injected humour or randomised words which dont look even believable genera on the Internet tend to repeat predefined chunks necessary making Internet.Diffrent Websites
Get in Touch
- 2901 Marmora Road, Glassgow,
Seattle, WA 98122-1090 - (088) -234 -456 -7890
- (088) -234 -456 -7890
- info@yourdomain.com
- http://domainname.com
- marta.filizola@mssm.edu
- + 1 212-659-8690
Molecular Modeling and Enhanced Molecular Dynamics Simulations of GPCRs (Especially Opioid Receptors) and Other Membrane Proteins
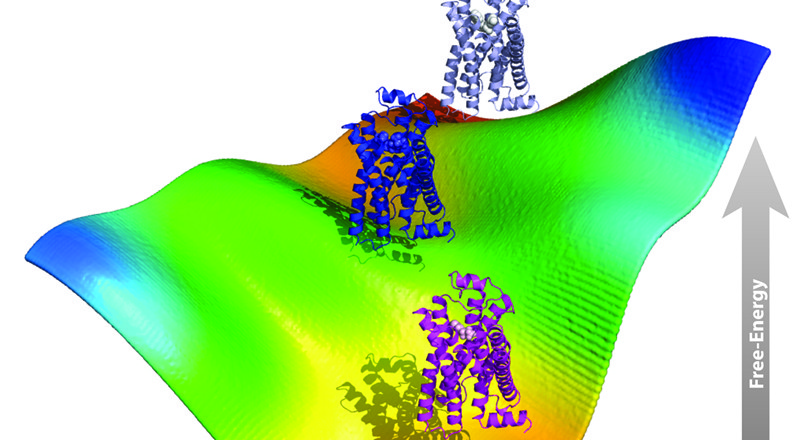
Among the milestones that we were able to accomplish under the auspices of continued NIH funding are the design, testing, and implementation of innovative computational strategies to build improved molecular models of G Protein-Coupled Receptors (GPCRs) and to study, more efficiently, the conformational plasticity and dynamical nature of liganded or unliganded, single or interacting, receptors within their natural lipid environment. In particular, we pioneered the use of enhanced, molecular dynamics (MD)-based computational strategies in combination with either atomistic or coarse-grained (CG) system representations to improve dynamic molecular models of GPCR molecular recognition, activation, and oligomerization, with the ultimate goal of elucidating receptor allostery and functional selectivity for successful use in rational drug design. In particular, we were able: (i) to obtain reliable models of ligand-bound conformations of GPCRs that do not require very long and computationally inefficient standard MD simulations, (ii) to establish a possible molecular basis for the functional selectivity of GPCRs through the prediction of ligand-specific conformations, and (iii) to advance current understanding of the role of oligomerization in receptor function through the generation of novel testable hypotheses of specific mutations that could eventually be used to modulate receptor function (see section 2, below). While the specific computational methods we explored are not new from an algorithmic standpoint, we developed some distinctive combinations of such methods, and showed in recent publications that they are indeed able to generate ligand-specific conformations of both isolated and interacting, inactive and active, GPCRs that are consistent with experimental data. Notably, the methodologies we developed represent fundamental tools that can be generalized to other transmembrane receptors, as well as broadly to other proteins.
Among the various GPCRs, we have devoted special attention to opioid receptors, which are important drug targets for pain management, drug abuse/addiction, and mood disorders. We have a long history of work on these GPCR subtypes, having contributed over the past 20 years to several structural and mechanistic insights into their pharmacology and signaling. Most recently, we have focused on their dynamics, seeking answers to questions like: How does an opioid drug bind to his receptor? How can sodium ions modulate opioid receptor activity? What are the likely interfaces of opioid receptor homodimers? What are the thermodynamic and kinetic elements of ligand binding and functional selectivity?
Representative Publications
Provasi, D., Bortolato A., Filizola, M. “Exploring Molecular Mechanisms of Ligand Recognition by Opioid Receptors with Metadynamics.” Biochemistry (2009) 48 (42): 10020-10029.
Filizola M. & Devi, L.A. “How opioid drugs bind to receptors” Nature (2012) 485, 314-7.
Johnston, J.M., Aburi, M., Provasi, D., Bortolato, A., Urizar, E., Lambert, N.A., Javitch, J.A., Filizola, M. “Making Structural Sense of Dimerization Interfaces of Delta Opioid Receptor Homodimers”, Biochemistry (2011) 50(10):1682-1690
Shang, Y., LeRouzic, V., Schneider, S., Bisignano, P., Pasternak, G.W., Filizola, M. “Mechanistic Insights into the Allosteric Modulation of Opioid Receptors by Sodium Ions” Biochemistry (2014) 53(31):5140-9.
Mobarec, J.C., Sanchez, R., and Filizola, M. “Modern Homology Modeling of G-Protein Coupled Receptors: Which Structural Template to Use?” Journal of Medicinal Chemistry (2009) 52 (16), 5207-5216.
Provasi, D., Camacho-Artacho, M., Negri, A., Mobarec, J.C., Filizola, M. “Ligand-Induced Modulation of the Free-Energy Landscape of G Protein-Coupled Receptors Explored by Adaptive Biasing Techniques.” PLOS Computational Biology (2011) 7(10):e1002193.
Provasi, D. & Filizola, M. “Putative Active States of a Prototypic G-Protein Coupled Receptor from Biased Molecular Dynamics.”Biophysical Journal (2010) 19(10):2347-2355.
Schneider, S., Provasi, D. & Filizola, M. “The Dynamic Process of Drug-GPCR Binding at Either Orthosteric or Allosteric Sites Evaluated by Metadynamics” Methods in Molecular Biology (2015) 1335:277-294.
Provasi, D., Boz, M.B., Johnston, J.M., Filizola, M. “Preferred Supramolecular Organization and Dimer Interfaces of Opioid Receptors from Simulated Self-Association” PLOS Computational Biology (2015) Mar 30;11(3):e1004148
Shang, Y., Yeatman, H.R., Provasi, D., Alt, A., Christopoulos, A., Canals, M., and Filizola, M. “Proposed Mode of Binding and Action of Positive Allosteric Modulators at Opioid Receptors” (2016) ACS Chemical Biology; 11(5):1220-9
Schneider, S., Provasi, D., Filizola, M. “How Oliceridine (TRV-130) Binds and Stabilizes a μ-Opioid Receptor Conformational State that Selectively Triggers G Protein-Signaling Pathways.” (2016) Biochemistry55(46):6456-6466
Marino, K., Prada-Gracia, D., Provasi, D., Filizola, M. “Impact of Lipid Composition and Receptor Conformation on the Spatio-Temporal Organization of mu-Opioid Receptors in a Multi-component Plasma Membrane Model” (2016) PLOS Computational Biology 12(12):e1005240
Kapoor, A., Martinez-Rosell, Provasi, D., de Fabritiis, G., and Filizola, M. “Dynamic and Kinetic Elements of µ-Opioid Receptor Functional Selectivity” Nature – Scientific Reports, (2017) 7(1):11255.
Marino, K.A. & Filizola, M. “Investigating Small-Molecule Ligand Binding to G Protein-Coupled Receptors with Biased or Unbiased Molecular Dynamics Simulations” (2018) “Methods in Molecular Biology”, 1705: 351-364.
Filizola, M. “Insights from Molecular Dynamics Simulations to Exploit New Trends for the Development of Improved Opioid Drugs” (2018) Neuroscience Letters, Feb 18. pii: S0304-3940(18)30119-8.
Meral, D., Provasi, D., Prada-Gracia, D., Möller, J., Marino, K., Lohse, M.J., and Filizola, M. “Molecular details of dimerization kinetics reveal negligible populations of transient µ-opioid receptor homodimers at physiological concentrations” (2018) Scientific Reports 8(1):7705
Walsh, S., Mathiasen, S., Christensen, S.M., Fay, J.F., King, C., Provasi, D., Borrero, E., Rasmussen, S.G.F., Fung, J.J., Filizola, M., Hristova, K., Kobilka, B., Farrens, D.L., Stamou, D. “Single proteoliposome high content analysis reveals differences in the homo-oligomerization of GPCRs” (2018) Biophysical Journal 115(2):300-312.
Meral, D. , Provasi, D., Filizola, M. “An Efficient Strategy to Estimate Thermodynamics and Kinetics of G Protein-Coupled Receptor Activation Using Metadynamics and Maximum Caliber” bioRxiv; https://doi.org/10.1101/367888; (2018) Journal of Chemical Physics 149(22):224101
Hu, X., Wang, Y., Hunkele, A., Provasi, D., Pasternak, G.W., Filizola, M. “Kinetic and thermodynamic insights into sodium ion translocation through the μ-opioid receptor from molecular dynamics and machine learning analysis” (2019) PLOS Computational Biology, 15(1):e1006689
Ribeiro, JML & Filizola, M. “Allostery in G Protein-Coupled Receptors Investigated by Molecular Dynamics Simulations” (2019) Current Opinion in Structural Biology Volume 55: 121-128
Ribeiro, JML & Filizola, M. “Insights from Molecular Dynamics Simulations of a Number of G-Protein Coupled Receptor Targets for the Treatment of Pain and Opioid Use Disorders” Frontiers in Molecular Neuroscience (2019) 12:207
Hu, X., Provasi, D., Filizola, M. “Mechanism of μ-Opioid Receptor-Magnesium Interaction and Positive Allosteric Modulation.” bioRxiv 689612; doi: https://doi.org/10.1101/689612; (2019) Biophysical Journal, pii: S0006-3495(19)30854-9
Kapoor, A., Provasi, D., Filizola, M. “Atomic-Level Characterization of the Methadone-Stabilized Active Conformation of µ-Opioid Receptor” (2020) Molecular Pharmacology, mol.119.119339.
Ribeiro Lamim, J.M., Provasi, D., Filizola, M. “A combination of machine learning and infrequent metadynamics to efficiently predict kinetic rates, transition states, and molecular determinants of drug dissociation from G protein-coupled receptors” Journal of Chemical Physics (2020) 153(12):124105
Zhou, Y., Ramsey, S., Provasi, D., El Daibani, A., Appourchaux, K., Chakraborty, S., Kapoor, A., Che, T., Mazumdar, S., Filizola, M. “Predicted Mode of Binding to and Allosteric Modulation of the m-Opioid Receptor by Kratom’s Alkaloids with Reported Antinociception in Vivo” Biochemistry (2020) Dec 4. doi: 10.1021/acs.biochem.0c00658
Pryce, K.D., Kang, H.J., Sakloth, F., Liu, Y., Khan, S., Toth, K., Kapoor, A., Nicolais, A., Che, T., Qin, L., Bertherat, F., Kaniskan, H.U., Jin, J., Cameron, M.D.,Roth, B.L., Zachariou, V., and Filizola, M.“Positive Allosteric Modulators of the m-Opioid Receptor Enhance the Antinociceptive Efficacy of Opioids but not their Adverse Effects” (2021) Neuropharmacology Sep 1;195:108673
Chakraborty, S., Diberto, J., Faouzi, A., Bernhard, S., Gutridge, A., Ramsey, S., Zhou, Y., Provasi, D., Nuthikattu, N., Jilakia, R., Nelson, M.N.F., Asher, W.B., Eans, S. O., Wilson, L.L., Chintala, S.M., Filizola, M., van Rijn, R.M., Margolis, E.B., Roth, B.L., McLaughlin, J.P., Che, T., Sames, D., Javitch, J.A., Majumdar, S. “A novel mitragynine analog with low efficacy mu-opioid receptor agonism displays antinociception with attenuated adverse effects” (2021) Journal of Medicinal Chemistry Sep 23;64(18):13873-13892.
Salas-Estrada, L., Fiorillo, B., Filizola, M. “Metadynamics Simulations Leveraged by Statistical Analyses and Artificial Intelligence-Based Tools to Inform the Discovery of G Protein-Coupled Receptor Ligands” (2022) Frontiers in Endocrinology, Section Cellular Endocrinology; 13:1099715.
Ahn, D., Provasi, D., Duc, N.M., Xu, J., Salas-Estrada, L., Spasic,, Yun, M.W., Kang, J., Gim, D., Lee, J., Du, Y., Filizola, M., Chung, K.Y. “Gαs slow conformational transition upon GTP binding and a novel Gαs regulator” bioRxiv (2022) doi: https://doi.org/10.1101/2022.10.10.511514; (2023); iScience Apr 8;26(5):106603.
 Creative Style
Creative Style Portfolio Style
Portfolio Style One Page Style
One Page Style Landing Page
Landing Page